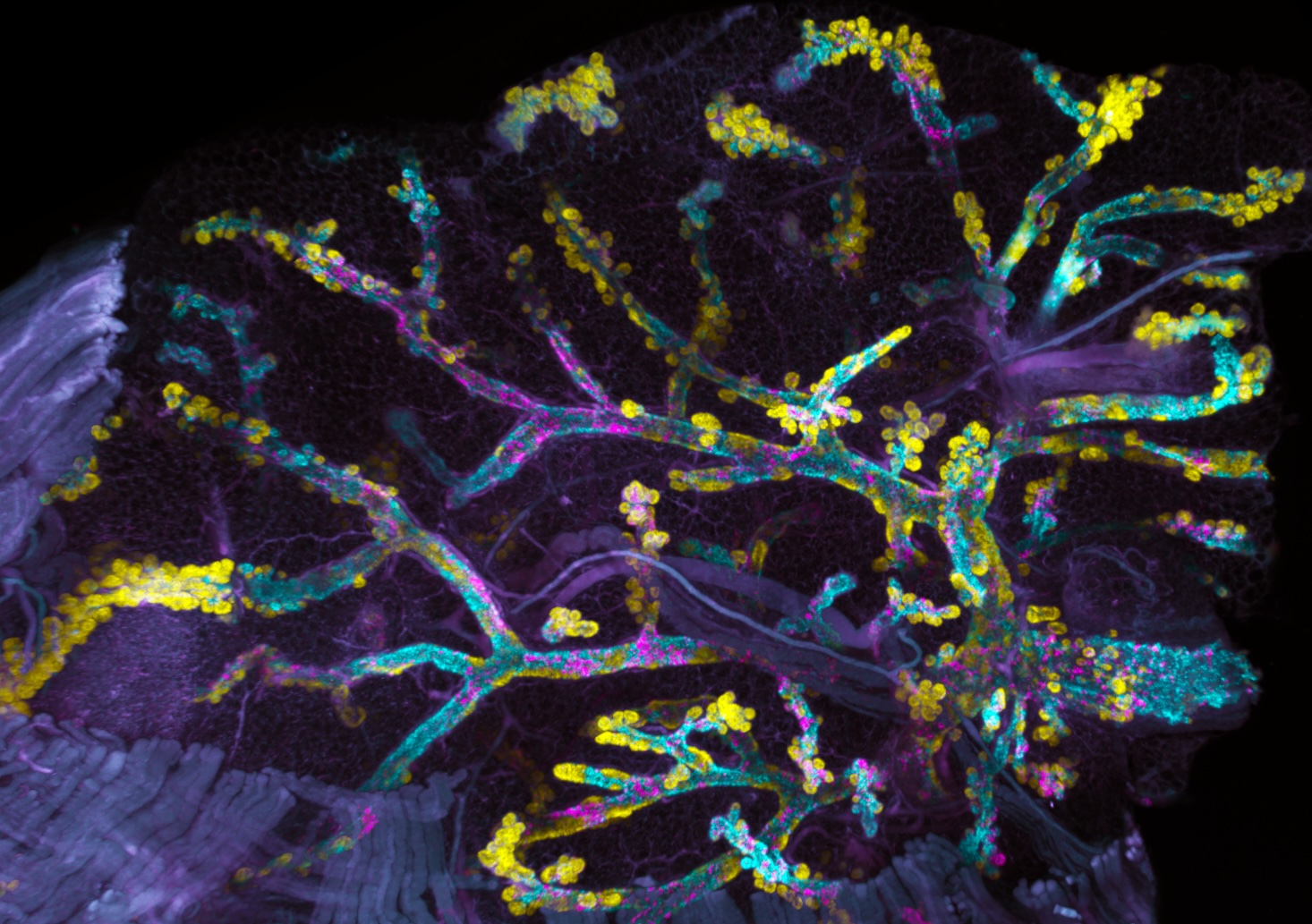
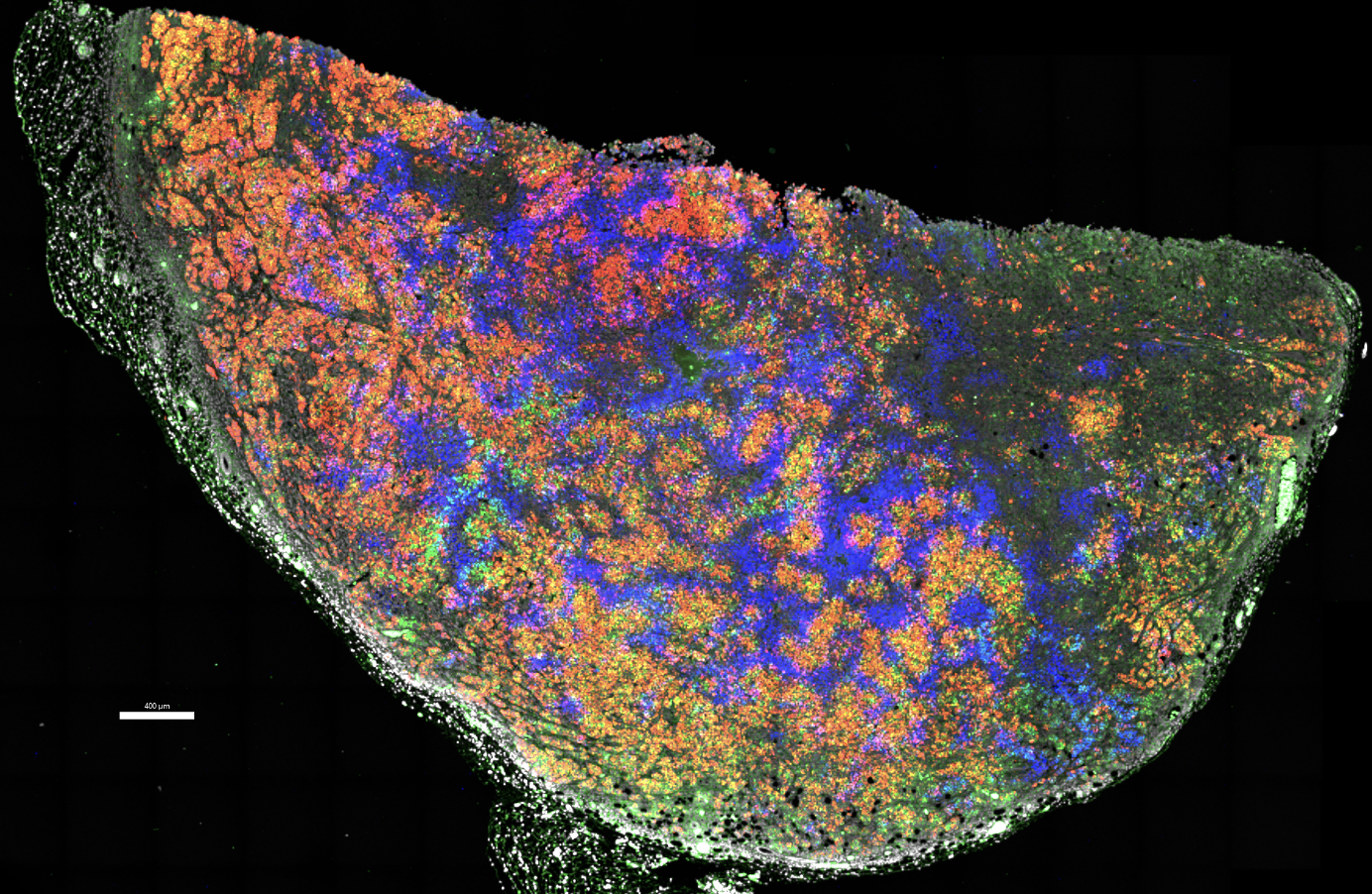
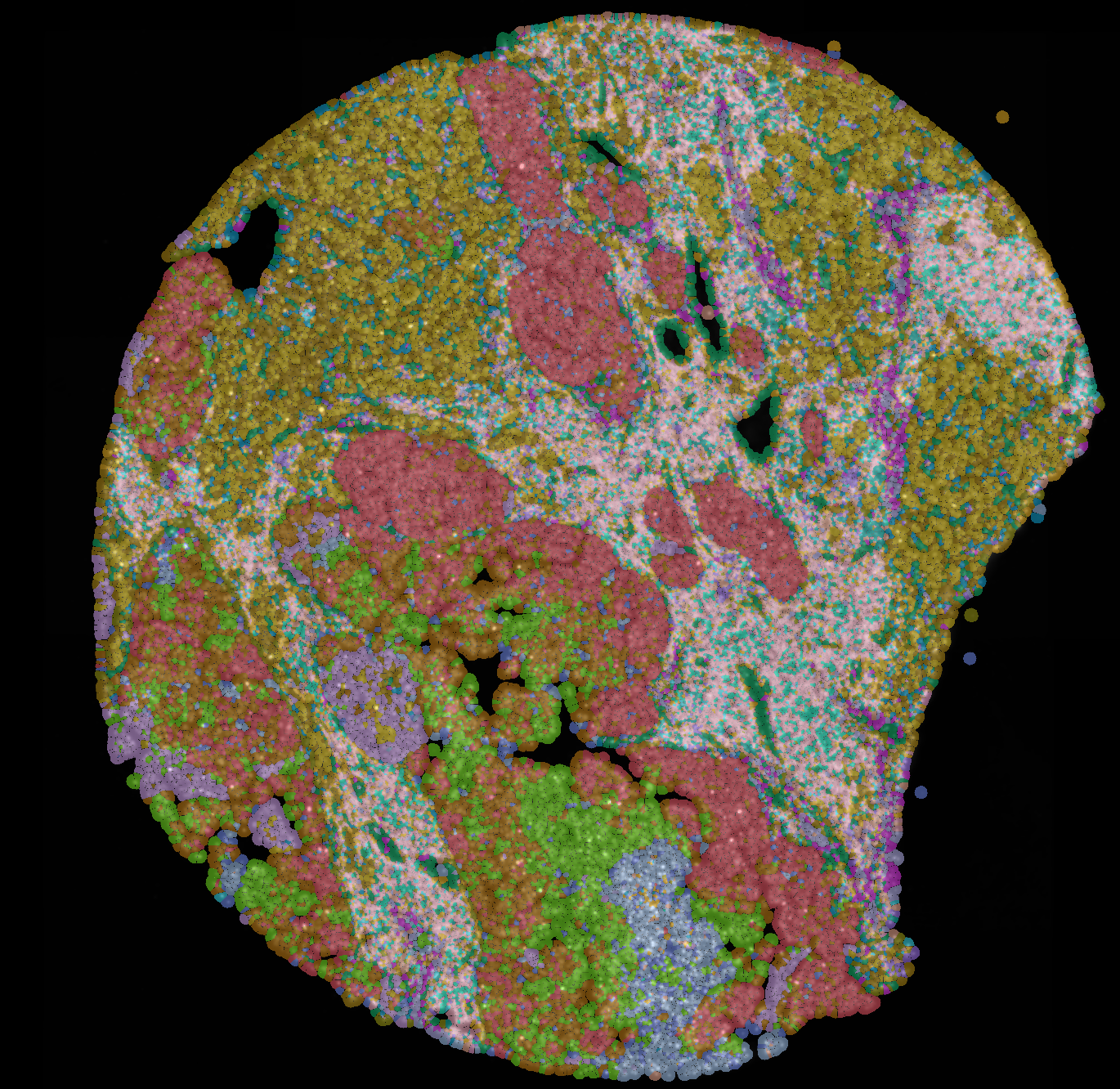
Overview
Our primary focus is to determine how cancer cells and their ecosystem adapt during the occult phase of tumorigenesis. We utilize Cancer rainbow models (Crainbow) and hyperspectral techniques for exploring tumor initiation with unprecedented resolution in space and time. We are highly interdisciplinary in our approach and utilize systems biology, spatial single cell analyses, and mathematical modeling.
Lab Projects

Somatic mutations drive phenotypic changes in cellular behaviors. Mutations to cancer causing tumor driver genes may be acquired years, if not decades before clinical diagnosis of cancer. What happens to the cell and how collective tissue behaviors shape the trajectory of a lethal cancer during a prolonged occult tumorigenic phase is a great mystery. Our lab has developed a set of mouse tools for illuminating the earliest initiation phase of cancer. Our primary objective is to map the cellular events and biochemical interactions at work during the initiation phase of cancer in order to discover strategies for the prevention or treatment of lethal cancers. We use the mouse to fluorescently barcode somatic mutations and then observe cell competition, cell adaptation, and tumorigenesis in vivo. Our current focus is three-fold:
Building fluorescent-barcoded mice for visualizing cancer initiation
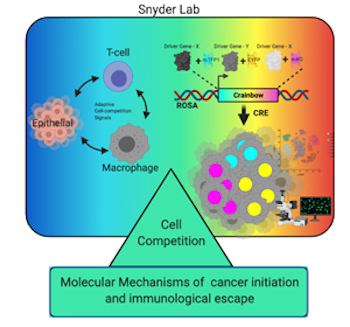
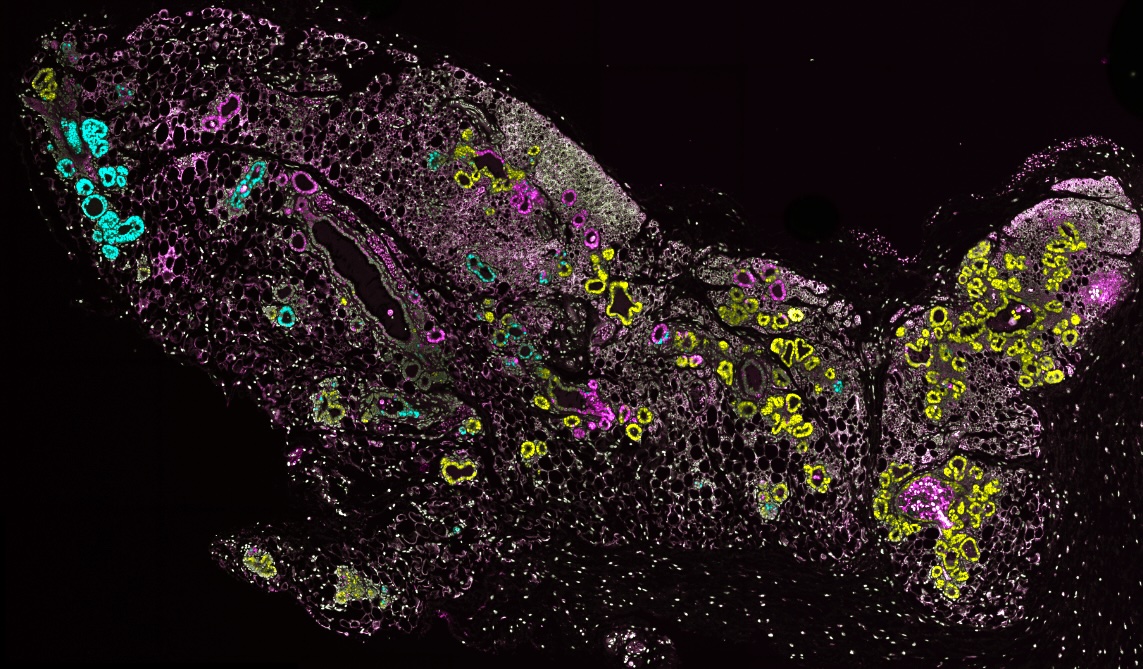
We have developed Cancer rainbow mice (Crainbow) in order to visualize the effects on cells immediately after oncogene expression. For a full description of Crainbow see our publication in Nature Communications or read our blog article posted to “Behind the Paper” on Nature Cancer Community.
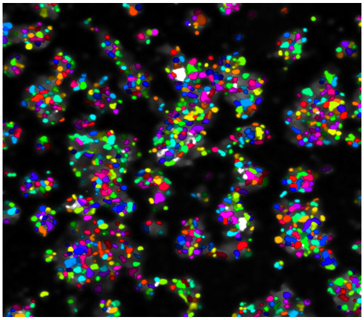
Our goal is to use Cancer rainbow mice for measuring cell competition (or maybe even cooperation) during the occult tumorigenic phase of cancer. All Crainbow models (8 and counting) are immune intact and enable us to study emergent cell behaviors in the presence of a normal immune response. We are also working toward imaging modalities and registration techniques for overlaying of histopathology and lineage tracing data (i.e. “TruRegistry”).
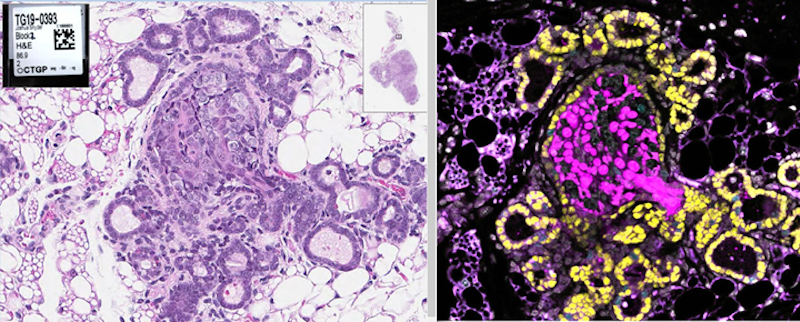
Intestinal stem cell competition in vivo and ex vivo
How do somatic mutations spread throughout the intestine? We recently showed that this can happen one of two ways, either during a postnatal growth phase of the intestine or through oncogenesis of microenvironmental cues (i.e. Rspondins). Read more here and see one of our favorite images from the study below.
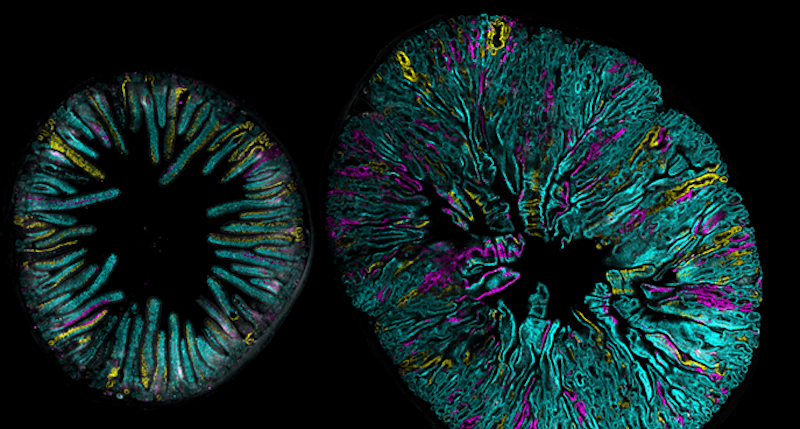
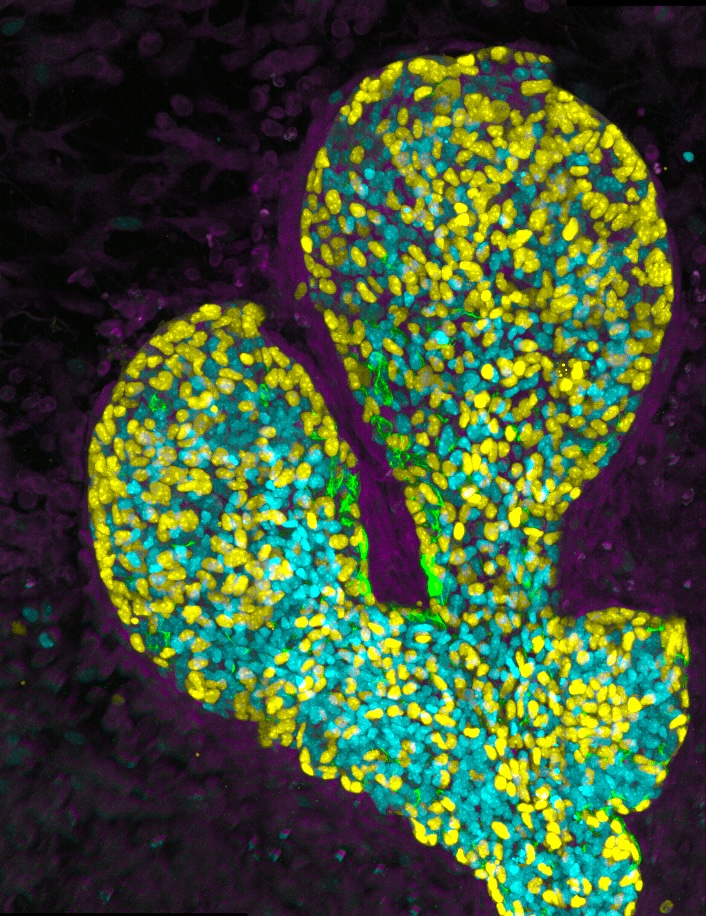
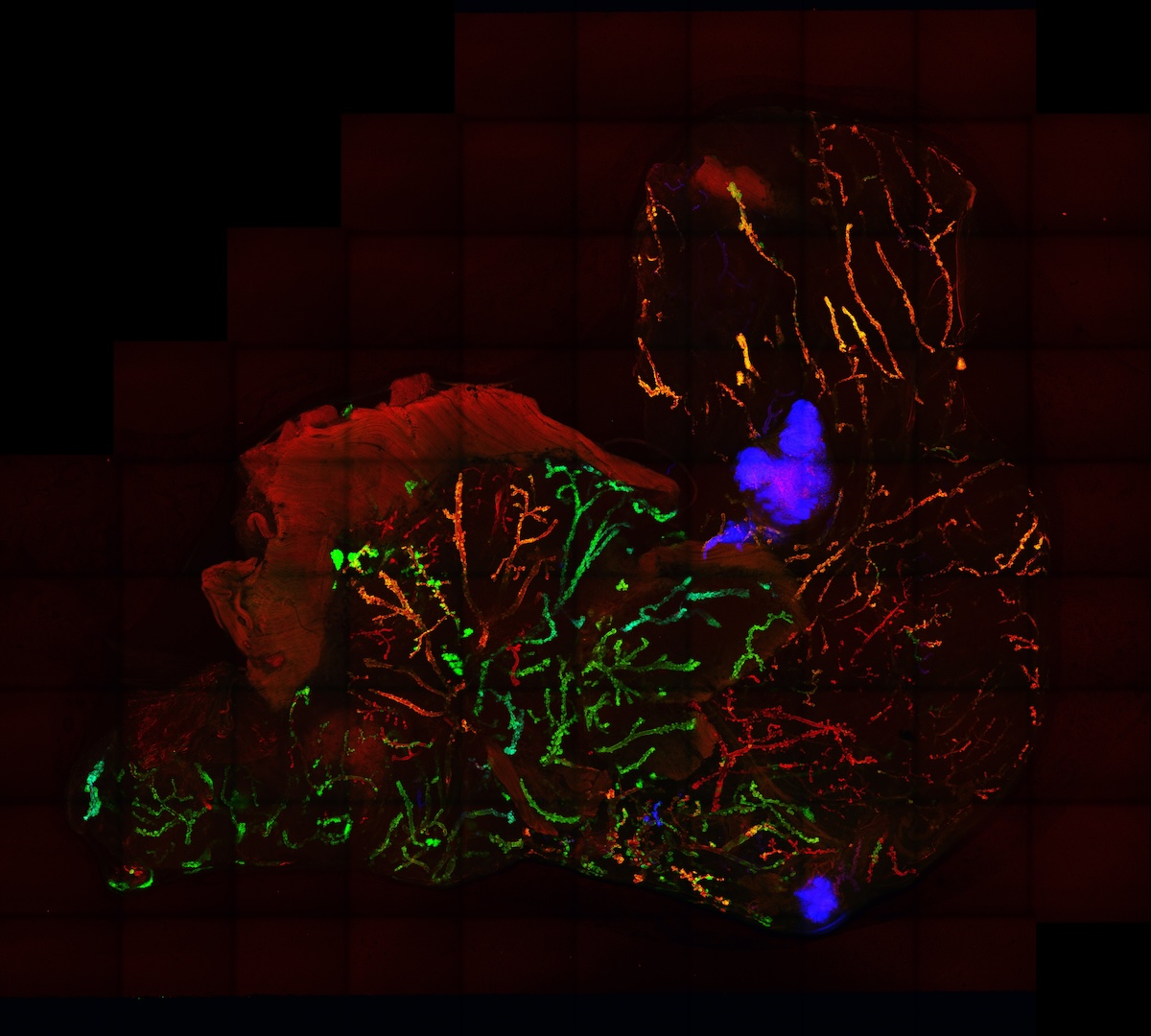
Imaging three-dimensional cellular networks during cancer progression
Our lab is recently interested in mapping the cellular behaviors and networks present during the initiation of breast cancer. New protocols for clarifying whole-organoids are being used in our Crainbow mice so that we can three-dimensionally map cell competition during the developmental phase of the mammary gland. Here, is one such movie of a three-dimensional projection of premalignant growth in the mammary gland – stay tuned, more progress in the mammary gland is coming soon!
The Snyder lab is part of the Department of Surgery’s Division of Surgical Sciences and affiliated with the Center for Applied Therapeutics. We have a secondary affiliation with the Department of Cell Biology and are also members of the Development and Stem Cell Biology (DSCB) and Cell and Molecular Biology (CMB) training program, as well as the Duke Cancer Institute.
Selected Achievements/Newsworthy
- Crainbow in the news as a "Biomedical Picture of the Day"
- Molecular Cancer Research Cover

Advanced Training
Routine technologies and tools used by the Snyder lab include hyperspectral fluorescent microscopy, whole-organ imaging, genetic engineering, stem cell biology, mouse models, bioinformatics, scRNAseq, lineage tracing, and spatial transcriptomics. The Snyder lab has hyperspectral confocal imaging, hyperspectral high-content live imaging, and fluorescent dissecting microscopes.
Contact Us
Are you interested in imaging, mouse genetics, and discovering how cancer cells are born? We need cancer biologists, molecular biologists, and bioinformaticians interested in all things imaging/big data. We are always looking to grow our team, including a post-doctoral fellowship, graduate student training, technical position, or undergraduate research opportunity. Send us an email.
Collaborators
Want to collaborate or have a reagent request? We like to share and support team science. Send us an email.
Publications and Funded Projects
View Dr. Snyder's profile to see his publications and funded projects.
Lab Members
Principal Investigator
PI of the Snyder Lab (Est. 2017). Josh received his PhD from the University of Pittsburgh, where he studied interactions of lung stem cells and the immune system during wound repair. As a post-doctoral fellow, he studied Lgr5 signaling and developed the first Cancer rainbow mouse (Crainbow) line to study cell-fitness effects of Lgr5 mutations. Crainbow mice brought a new direction to the lab, and we are now primarily focused on understanding the cellular and molecular principles underlying tumor initiation and growth. Follow Josh on Twitter (@Dnace128) for up-to-date news in the lab. Outside the lab you will find Josh enjoying the outdoors with his family.
Joshua Ginzel, PhD student

The majority of cancer mitigation strategies seek to detect and treat early stage disease; yet, in many cancers this strategy has been largely ineffective at reducing lethal, metastatic disease. Growing evidence suggests that the majority of cancer development occurs long before current detection methods are able to intercept the nascent lesion. My research interests focus on studying this invisible phase of tumorigenesis using systems biology approaches in mouse models of cancer.
Joe Fernandes, PhD student

My thesis work is focused on understanding how expression of oncogenic isoforms of HER2+ on breast cancer epithelial cells can lead to drastically different phenotypes such as difference in cell growth, motility, signaling, intratumoral competition, and drug resistance. For these dynamics, I use fluorescently barcode HER2+ mammary epithelial cell lines derived from our HER2 Crainbow mouse and leverage several in vitro and in vivo assay platforms along with mathematical modeling. Overall, I hope my work can inform further investigation into why HER2+ breast cancers recur and therapeutic approaches to prevent recurrence. Outside of lab, my interests include soccer, food, dogs, and being outdoors.
Alejandra Patino, MD-PhD student

My main research interests encompass the intersection of cellular plasticity and clonal evolution throughout the malignancy cascade. To study how cellular plasticity arises and contributes to invasion or treatment resistance, we need to reliably trace different clones in various contexts. During my PhD I have worked on identifying the maximum number of spectrally resolvable fluorescent proteins for in-vivo applications. Utilizing this methodology, my remaining thesis work will focus on understanding how early Epithelial to Mesenchymal Transition (EMT) states arise and contribute to invasion and treatment resistance. Outside of lab I enjoy exploring the Durham food scene, pickleball, spin, and cooking with friends.
Natalie Thomas, PhD student

My thesis project focuses on the intersection of development and oncogenesis in the context of the mammary epithelium. Our lab has identified the nuclear alarmin IL33 as a target for further study due to its unusual expression patterns in the mammary epithelium, exhibiting high levels of expression during development and cancer, but not at rest. To examine this relationship, I use a combination of in vitro and in vivo models to better understand the necessity and mechanism of action of IL33 in the mammary epithelium in both contexts. This combinatorial approach utilizes various engineered mouse models, molecular assays, single-cell and spatial transcriptomics, and confocal microscopy. I hope that my research can provide the groundwork for future breast cancer therapies and improve the understanding of the relationship between cancer and development. Outside the lab, I enjoy live music, exploring the outdoors, and trying out new recipes.
Hillary Zawada, research technician

I have a background in evolutionary anthropology with an interest in genetics, epigenetics, and evolutionary biology. In our lab, I’m interested in factors that influence cellular competition and dynamics such as oncogene-induced senescence (OIS), gene regulation, cellular signaling, and relationship with the immune system. I manage over 50 genetic strains for our mouse models and assist with research wherever I can. Outside of the lab I enjoy reading, hiking, going to the beach, and playing videogames.
Henry Chapman, clinical data manager

My work focuses on aiding research collaborators with their computational needs, specializing in microscopy image analysis and segmentation. Using Cellpose, an open-source image segmentation algorithm, I segment images from both confocal and bright field microscopy and then analyze the cells present to determine things such as their fluorescent protein tag or tumor genotype. Most recently I have been working towards implementing a machine learning algorithm to aid in the analysis of the cellular data. Previously I have written code to simulate and predict Her2+ tumor progression using differential equations and data sampling to fit the model parameters to best describe the observed data. Outside of the lab I like to listen to vinyl records, play pool, and go rock hounding.
Lab Photos